
Source: Pexels
Introduction
Suppose that in an office, several letters are being issued in each and every member’s name separately from a single source for any purpose. Now, only those letters which bear the correct postal code or address of the receiver will reach their target destination. But those with an error in the postal code or address will not reach the target receiver. Likewise, all proteins are synthesized on ribosomes in the cytosol regardless of their target site of action. So, naturally, a question will arise – How all proteins are directed to their individual final cellular destinations appropriately? Because proteins don’t know where to go and work. Does there exist any sort of postal code even in proteins that are playing a pivotal role? So, let’s understand this complex and fascinating process, particularly for eukaryotic cells.
The signal sequence
In eukaryotic cells, all proteins start their synthesis in the cytosol but not all of them complete the job here. It depends on the primary cellular destination of the proteins. Proteins destined for secretion, integration in the plasma membrane or lysosomal inclusion, are all directed to the endoplasmic reticulum. Some proteins are destined for the nucleus, the mitochondria and the chloroplasts. Remaining ones which are destined for cytosol simply remain where they are synthesized. In 1970, scientist Gunter Blobel and his colleagues first postulated that this targeting of proteins to their cellular destinations entirely depends on a short stretch of amino acids mostly in the N-terminal of the nascent polypeptide which is called Signal Sequence. It directs a protein to its appropriate primary cellular location and in most cases, is cleaved after its job is done. Thus, it acts basically as a postal code of the proteins. It is confirmed by fusing the signal sequence of one protein in a second one and showing that the second protein is heading towards the destination of the first protein. They vary in length but have the following listed features in common:
- about 10 to 15 hydrophobic amino acid residues
- one or more positively charged residues at the beginning
- a relatively polar short sequence of amino acids at the carboxyl terminus
- a cleavage site

Let’s focus on the first category of targeting i.e. Endoplasmic Reticulum. Shall we?
Journey towards endoplasmic reticulum
The best-characterized targeting system occurs in the Endoplasmic Reticulum (ER). Most proteins: membrane, secretory and lysosomal, have a kind of signal sequence in their amino-terminal which targets them for localization in the ER.
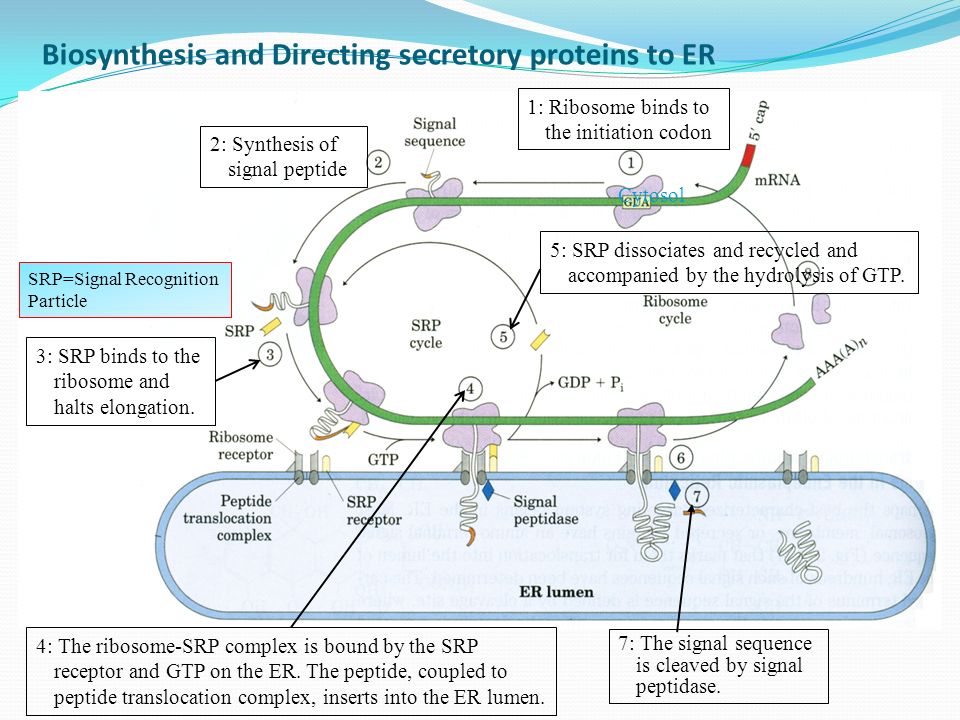
- Protein synthesis starts in free ribosomes of the cytosol. As mentioned before, due to the presence of signal sequence in the amino terminal, it is synthesized first.
- When it emerges out of the ribosome, the signal sequence along with ribosome is recognized and bound by a Signal Recognition Particle (SRP).
- SRP halts elongation of the polypeptide by restricting the passage of translation factors to the A site of the ribosome by binding to GTP.
- Then, the GTP bound SRP directs the ribosome and the incomplete polypeptide to the SRP receptors present on ER.
- The nascent polypeptide then is channelized to peptide translocation complex in the ER where it interacts with the ribosomes bound to ER.
- SRP dissociates by GTP hydrolysis and translation continues now.
- Finally, when the complete protein is synthesized, the signal sequence is cleaved off by signal peptidases within the ER.
Post-translational modifications in ER and Golgi apparatus
The signal peptide directs the proteins to the ER. In the ER, various protein folding, disulphide bond formation and other post-translation modifications begin. Glycosylation is an important modification that proteins undergo in ER to increase the proteomic diversity, wherein various carbohydrate moieties are attached to the proteins. Glycosylation helps in targeting proteins to the correct destination, cell-matrix adhesion and other signal transduction pathways. Our blood group antigens on RBCs are differentiated by the sugar residues they contain.
D-Glucose, D Galactose, D-Mannose, L-Fucose, N-Acetyl Glucosamine, N-Acetyl Galactosamine etc. are common carbohydrate moieties that participate in glycosylation. Most of the glycosylations which occur are N-linked (i.e. sugar moieties attached to the amino group of asparagine) or O-linked (i.e. sugar moieties attached to the hydroxyl group of serine or threonine). Besides that, additional modifications occur in the Golgi Apparatus before finally sending them to their final cellular destinations i.e. either cell or organelle membrane, extracellular matrix or lysosomes.
Mannose-6-phosphate: the lysosomal code
In the Golgi, after the proteins have been glycosylated, their final destination journey starts. In proteins destined for lysosomes, a mannose residue on the surface is phosphorylated forming mannose 6-phosphate, which is the structural signal.

The fancy story of the membrane proteins
Only signal peptide sequence is not enough for proteins to reach their final destination. It needs additional sequences which will guide them towards their destination from the ER or Golgi. Membrane proteins additionally contain some sequences in the middle which are called topogenic sequences (Stop Transfer Sequence and Signal Anchor Sequence). These determine the protein’s orientation in the cell membrane or membranes of ER, Golgi etc. The most important of them is called Signal Anchor Sequence.
It is a 22-25 long hydrophobic amino acid chain that predominantly forms alpha-helix with 3 positively charged residues either at its left or right. This determines their orientation in the ER membrane. There are three scenarios:
- Positively charged amino acids located on N-terminal of hydrophobic sequence: protein will be oriented transmembrane with its N terminus into the cytosol and C terminus into the lumen of ER as positive charges have to stay cytosolic.
- Positively charged amino acids are located on C-terminal of hydrophobic sequence: protein will be oriented transmembrane with its N terminus into the lumen and C terminus into the cytosol.
- For multipass protein, the scenario becomes complex because here, the position of N terminus will depend on orientation of first signal sequence. The position of C terminus will depend on the total number of stop transfer and signal anchor sequences. If it is even, then C terminus will end in same side of N terminus. But if it is odd, the C terminus will end in the opposite side.

Most proteins leave the ER, transported in vesicles to the Golgi and from there they are either secreted, or travel to the cell membrane, or bound for lysosomes. Likewise, for membrane proteins, secretory proteins must also contain some additional conserved sequences for their transport towards the extracellular matrix.
The right cargo, at the right place, at the right time: the introduction of SNARE hypothesis
So far, we have seen that the proteins are transported in the form of cargo to their proper destination with the help of some carriers called vesicles. Now there are various kinds of vesicles that transport their materials mainly proteins from one membrane component to another. It mainly does two things i.e. pinching off a membrane called vesicle budding with its necessary cargo and coat proteins and then fusion to its target membrane and delivery of its contents. So, as the heading suggests, for the right cargo to go to the right place, requires high specificity. Our main focus in this section will, therefore, be on the high specificity achieved in the process of membrane fusion and its mechanism.

A brief history
The story began almost 30-35 years ago when James Rothman (2013 Nobel Laureate in Medicine or Physiology) and his laboratory made the breakthrough that membrane fusion is blocked and the transport vesicles accumulate on the addition of a reagent i.e. N-ethylmaleimide (NEM). That means, there might be any N-ethylmaleimide-sensitive factor (NSF) that is necessary for membrane fusion. Their assumption was right, so they were successful in purifying NSF from crude cell extract and observed that when added back, it restores transport and fusion following NEM inactivation.
But there is a mystery again. NSF is a water-soluble ATPase that does not usually bind to membranes. How then a protein which does not bind to membranes is engaged in membrane fusion?
SNAP-SNARE complex
There must be some NSF attachment protein that binds NSF and recruit it to membranes specifically Golgi membranes which were later found and named SNAP (Soluble NSF attachment protein). SNAP binds to SNAREs (Soluble NSF Attachment Protein Receptors) which ultimately performs membrane fusion. But the main thing happened when they turned their attention to the brain tissue where three SNARE proteins, Synaptobrevin, SNAP 25 and Syntaxin are located out of which Synaptobrevin resided on the vesicle and rest two on the plasma membrane.
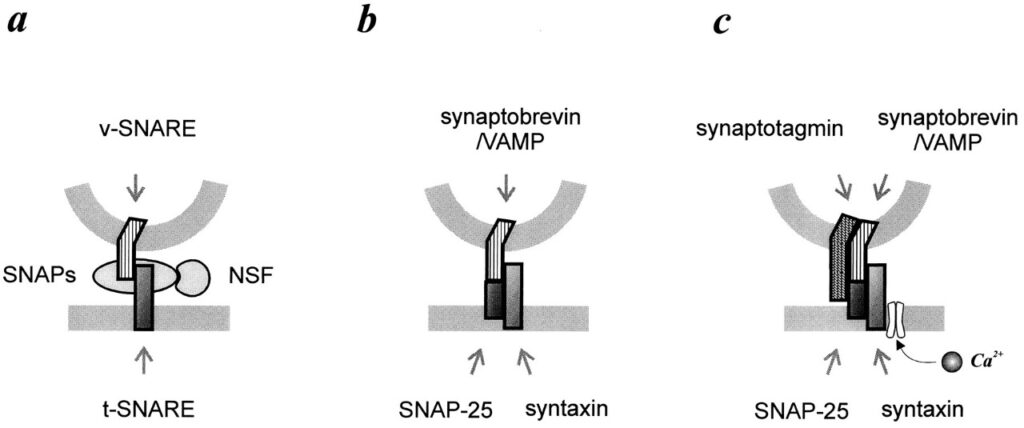
That’s how the SNARE hypothesis was proposed and later proved that – each of the vesicles and target membranes must be marked somehow to indicate which vesicle will fuse where. There must be some complementarity and matching between the markers such that only matching pairs bind with themselves and then only membrane fusion takes place. Here SNAREs are those vesicle and target markers termed as v-SNAREs and t-SNAREs.
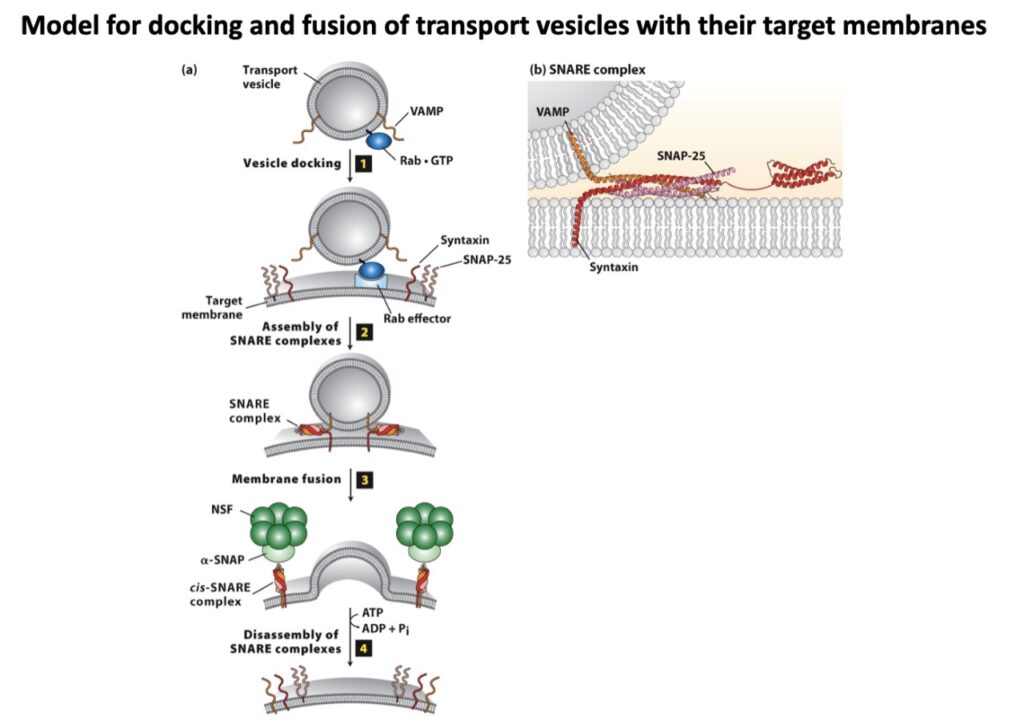
But you must be wondering where then NSF or SNAP functions if only SNAREs are required for fusion?
Actually NSF and SNAP help to disrupt SNARE complexes after fusion and help them to recycle for subsequent fusions. Thereby, they are also the main players along with SNAREs, without which membrane fusion will be stopped.
Conclusion
So far we have seen the role of signal peptide sequences and other additional sequences within a protein which are the sole determinants of a protein’s final destination. This article focuses on proteins that head towards ER i.e. a majority fraction of membrane, secretory and lysosomal proteins. In addition to that, there are proteins that travel towards mitochondria, nucleus, chloroplast etc. We have seen that every time signal peptides are cleaved after their job is done. But there are some proteins mostly nuclear proteins whose signal sequences are not cleaved and also some others which do not have any kind of signal sequence from the beginning. So the story is not over yet.
Questions to think about
I will conclude with some questions which need to be addressed in this regard:
- What signal sequence(s) is/are responsible for selectively phosphorylating mannose residues on the lysosomal proteins? Why is there a bias for phosphorylation on mannose especially?
- Is signal peptide mRNA transcribed/translated along with the protein coding region or is it transcribed/translated separately from a different region of DNA and then gets combined with the protein part?
- What is the exact reason of such conserved features of signal sequence?
- Which additional sequences or mechanisms determine whether a membrane protein will be inserted in cell membrane or an organelle’s membrane?
By Shraman Jana (2nd-year BS-MS student, IISER Kolkata)
References
- Purification of an N-ethyl maleimide sensitive protein catalysing vesicular transport – Block, Wieland and Rothman, 1988
- The Principle of Membrane Fusion in the Cell, Nobel Lecture by James Edward Rothman, 2013
- Lehninger’s Principle of Biochemistry, 7th edition
- Cell Biology 04: The Secretory Pathway- Harvard University Cell Biology Lecture Notes
About the author

I am a 2nd-year BS-MS undergrad in IISER Kolkata, KVPY fellow and recipient of NEET 2020 AIR 332. Being an aspired researcher by mind, I am a guy who has great affection towards biology and want to solve unknown and unanswered stuff in this subject. Apart from studies, I love reading storybooks, singing, play and watch football and cricket, play computer games and am a die-hard fan of FC Barcelona. I always believe in sharing knowledge and follows the quote- “If you want to shine like a sun, first burn like a sun.”
This is Shraman’s 2nd blog on TQR’s blogging platform. Find out his other blogs here!